Autophagy, the self-degradation mechanism that recycles damaged cellular components, is essential for maintaining cellular homeostasis. From early development to stress adaptation, this finely tuned process ensures cells survive unfavorable conditions and manage energy resources efficiently. Recent research reveals a fascinating evolutionary parallel in the transcriptional regulation of autophagy between humans and the fruit fly Drosophila melanogaster.

At the center of this discovery are two transcription factors: ZKSCAN3 in humans and M1BP in Drosophila. Despite arising in evolutionarily distant organisms, both play a conserved role as negative regulators of autophagy, controlling the expression of autophagy-related genes in the nucleus.
ZKSCAN3: A Master Transcriptional Repressor in Human Autophagy
ZKSCAN3 (Zinc finger with KRAB and SCAN domains 3) functions predominantly as a transcriptional repressor. Under nutrient-rich conditions, ZKSCAN3 remains in the nucleus and actively suppresses the transcription of autophagy and lysosomal genes. However, during starvation or cellular stress, ZKSCAN3 exits the nucleus, relieving this repression and allowing autophagy to proceed. This dynamic shuttling mechanism provides tight regulatory control over autophagy initiation and progression.

M1BP: The Drosophila Counterpart
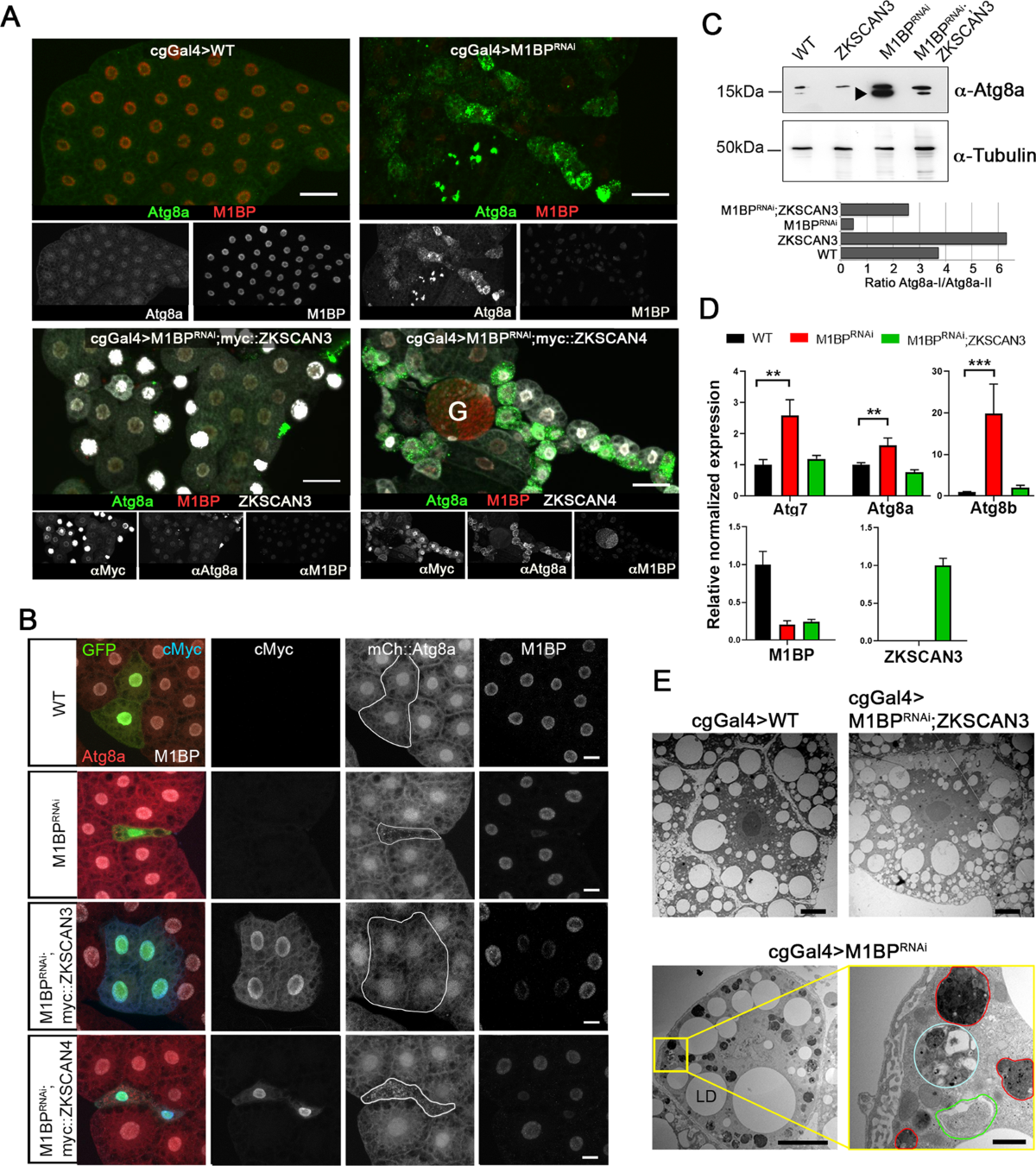
In Drosophila melanogaster, the M1BP (Motif 1 Binding Protein) plays an analogous role. Originally studied for its involvement in transcription pausing and promoter-proximal pausing in development, M1BP has been shown to repress genes involved in autophagy, mirroring the function of ZKSCAN3 in mammals. Loss of M1BP in fly cells induces upregulation of autophagy genes and leads to the formation of autophagosomes, the hallmark structures of active autophagy.
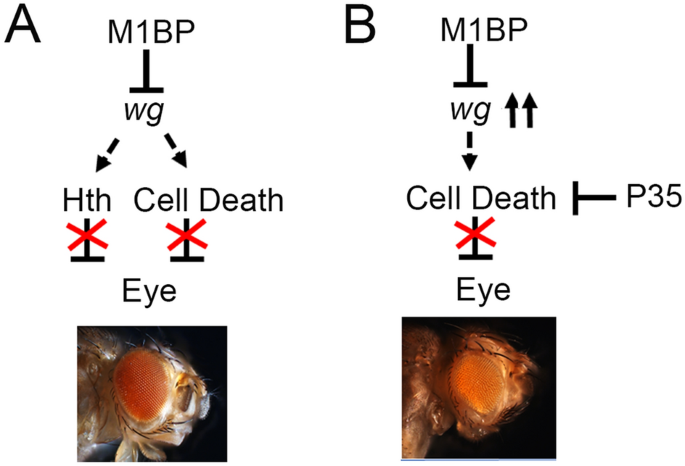
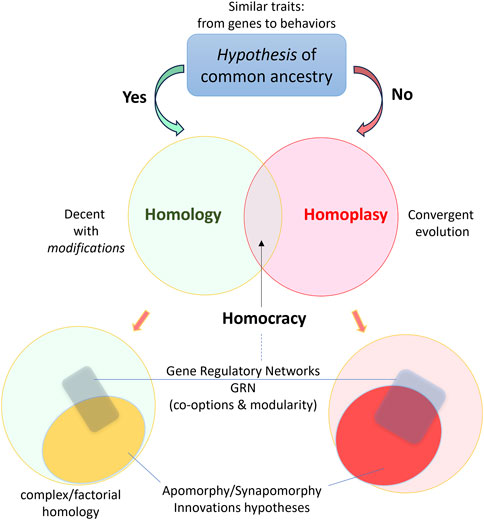
Functional Homology: Evolutionary Insights
Though ZKSCAN3 and M1BP are not sequence homologs, their functional homology—their ability to substitute for each other in cross-species experiments—highlights a remarkable example of convergent evolution in transcriptional control. In studies where M1BP is knocked down in Drosophila, introducing human ZKSCAN3 can restore repression of autophagy genes. This rescue effect strongly supports the idea that these proteins perform similar regulatory tasks across species, albeit through distinct molecular mechanisms.
Implications for biomedical research
Understanding the shared autophagy-regulating roles of ZKSCAN3 and M1BP opens new doors for biomedical research. Since dysregulated autophagy is implicated in cancer, neurodegeneration, and metabolic disorders, targeting transcriptional repressors like ZKSCAN3 could lead to novel therapeutic strategies. Furthermore, studying M1BP in Drosophila offers a genetically tractable model to investigate autophagy dynamics in vivo.
Conclusion
The discovery of functionally homologous transcription factors—ZKSCAN3 in humans and M1BP in Drosophila—adds a powerful dimension to our understanding of autophagy regulation. Despite evolutionary distance, nature has preserved core regulatory principles through functionally equivalent molecular players. Continued comparative studies will help us further unravel the complexities of autophagy and develop interventions to harness its therapeutic potential.